Unveiling the Lipid Recognition Mechanism of recluse spiders phospholipase D Toxin.
The venom of recluse spiders contains an enzyme known as phospholipase D (PLD) toxin. This toxin destroys the cell membranes of the preys by cleaving phospholipids, causing a set of symptoms called loxoscelism, which includes pain, swelling, and necrosis.
However, the mechanism by which the toxin recognizes specific phospholipids remains unknown. As different types of preys have membranes composed of different phospholipids, the lipid recognition mechanism of PLD toxins is relevant for their choice of prey.

Exploring recognition with simulations
Professor and Project Leader, Nathalie Reuter, Senior Engineer Emmanuel Moutoussamy and fellow researchers at the University of Bergen shed light on how PLD toxins recognise specific lipids. The researchers used molecular dynamics simulations which allow them to predict interactions between PLD toxins and various membrane compositions at the atomic level of detail. A preliminary analysis of the PLD toxins showed the presence of an aromatic cage (see figure) in the structure of some, but not all PLD toxins.
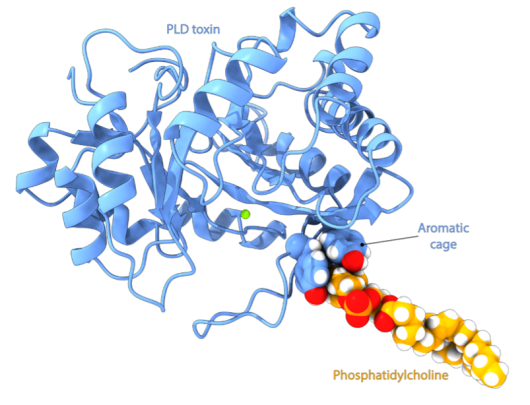
Such cages are known in other proteins to bind to specific phospholipids: phosphatidylcholine (PC). Our simulations revealed that this aromatic cage specifically binds to PC and explains why only certain PLD toxins can cleave PC. Given that phosphatidylcholine (PC) is the most prevalent phospholipid in mammalian membranes, our discovery provides an understanding of the specific targeting of this toxin towards mammals, which can be the prey of some recluse spiders. In addition, it enhances our general understanding of enzyme-lipid interactions and will be useful for the development of an anti-venom.
The role of high-performance computers (Betzy)
Accurate molecular dynamics simulations require the use of computers with ample processing power. These computers enable the simulation of large systems over time and the utilization of sophisticated algorithms for realistic simulations under various conditions. In this project, we utilized the computational capacity provided by Betzy, which required around 2 million CPU hours.